Vaccines gave us an antibody that vanquishes SARS-like Coronaviruses (including every variant of SARS-CoV-2 it was tested on)
The ultrapotent antibody SC27 could be a game-changer against SARS-CoV-2.
Cell Reports Medicine published a paper on August 20, 2024 that contains good news on the COVID-19 front (which I think we’re all due). Here’s the upshot: we know that hybrid immunity (the state of immunity resulting from a combination of vaccination and infection) offers the most robust protection from infection and downstream consequences of any option available (i.e., better than just infection or just vaccination), but we don’t fully understand why. A team of researchers found one potential reason: hybrid immunity endows people with exceptionally high quality antibodies. Among these was an antibody anointed with the catchy and euphonic1 sobriquet SC27, and it is VERY special. To understand why, we need to know a bit about antibodies in general, and a bit about antibodies to SARS-CoV-2:
Background
The paper intended to figure out what it is about hybrid immunity that makes it so effective in protecting people from SARS-CoV-2. To do this, it undertook a detailed examination of the antibody response to SARS-CoV-2 in infection, vaccination, and the combination. The really exciting part came from one specific antibody, SC27.
Antibodies to SARS-CoV-2’s spike protein are classified based on what part of the spike protein they target. Those that target the receptor binding domain (RBD), which is the part that engages with ACE2 (details on the structure of the RBD can be found in this footnote2) and lets the virus enter cells are further classified based on the part of the RBD they target as class 1/2/3/4 (classification of neutralizing antibodies is described in greater detail in this footnote3). Basically, the key differences between the classes are whether or not they bind within or outside the receptor-binding motif (RBM), and whether or not they bind the up or down conformation of the RBD (or both). To help us keep all of these straight, here is a cheat sheet from Gruell and colleagues (D is the most important part, but don’t feel like you need to memorize any of this- I honestly think the figure is very pretty and that was half my motivation for including it):

For a summary of the different conformations of SARS-CoV-2’s spike, see this footnote4.
In addition to these, since the publication of this classification scheme, Class 5 antibodies have also been described. These antibodies are noteworthy for their conserved target across SARS-like coronaviruses and are defined by the fact that they do not compete with ACE2 for binding spike or with the antibody CR3022. However, with the emergence of Omicron variants, many of these antibodies have substantially declined in their ability to neutralize SARS-CoV-2 and the potency of these antibodies varies significantly across specific antibodies.
Sometimes antibodies can fit into two of these classes at once e.g., class 1/4 antibodies, class 2/3 antibodies. Typically, this is based on which antibodies they compete with for binding, so, for example, an antibody that competes with class 1 and class 4 antibodies would be class 1/4 and an antibody that competes with both class 2 and class 3 would be class 2/3. In a very special case, an antibody (GAR12) that competes with class 1, 3, and 5 antibodies has been identified, which the authors refer to class 6 (I would call this a class 1/3/5 antibody); it neutralized all tested variants at a very low dose.
The Paper
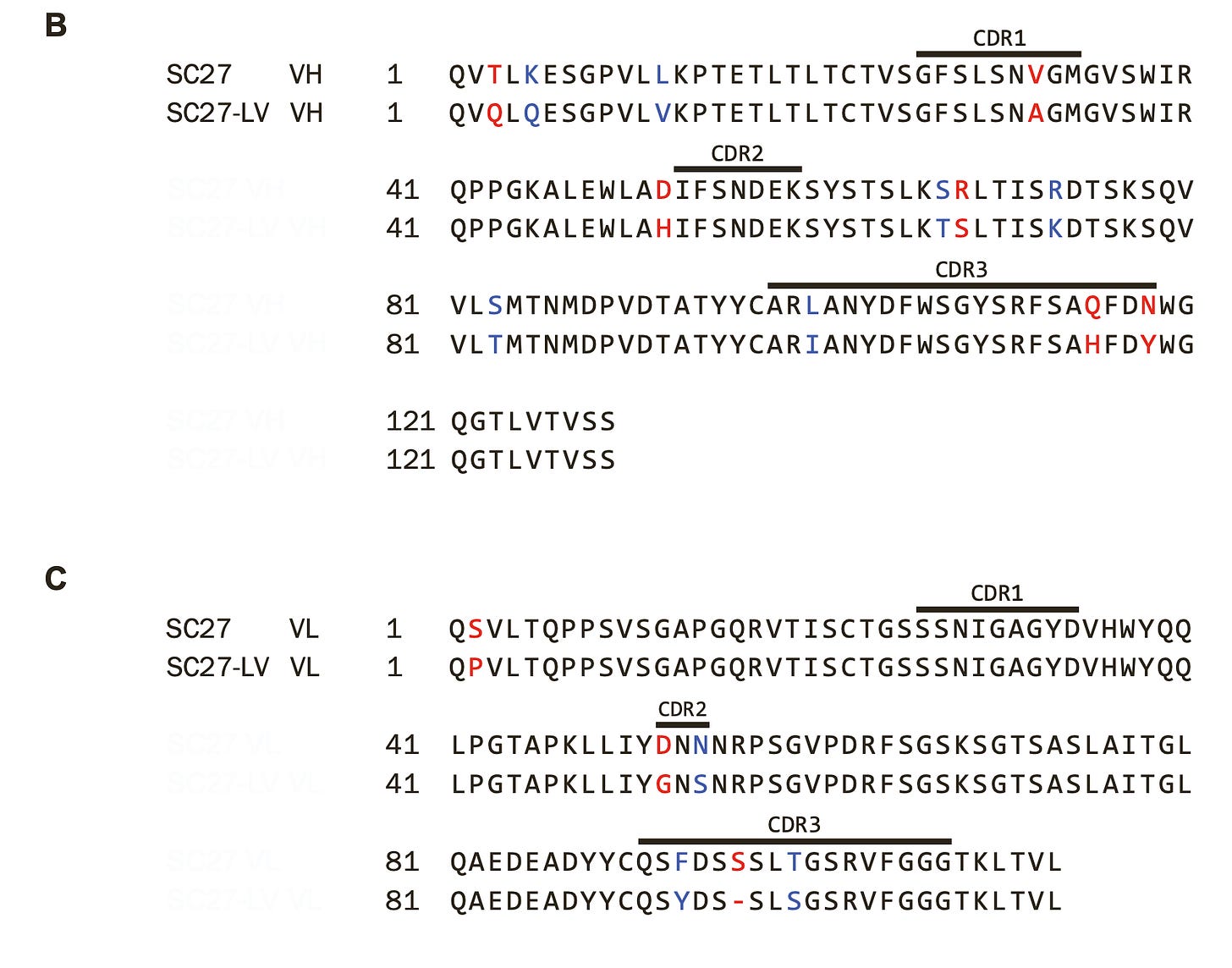
In this study, one of the donors received 2 doses of the Pfizer/BioNTech COVID-19 vaccine and then experienced an infection with a pre-Omicron variant. Before the breakthrough infection an antibody known as SC-27LV was present and it was pretty unremarkable:

The antibody could neutralize Pangolin-CoV, WIV1, and had some neutralization of D614G, Beta, Delta, and SARS-CoV-1 but essentially showed no ability to neutralize omicron variants or other coronaviruses. Fascinatingly, upon breakthrough infection of this vaccinee with a pre-Omicron variant, SC27-LV evolved into SC27 and this happened:
Suddenly, this previously unremarkable antibody becomes an extremely potent neutralizer of all SARS-like coronaviruses tested in vitro. And we even know which specific mutations are needed to go from SC27-LV to SC27:
A few other antibodies similar to SC27 were also found but they weren’t quite as good:
Notably, this also highlights that SC27 does not work on coronaviruses that are not SARS-like (sarbecoviruses; OC43 is an embecovirus and MERS-CoV is a merbecovirus). It is also noteworthy that SC27 is FAR more potent than the antibodies it is comparing with and it remains extremely potent even against BA.2.86 and JN.1. In fact, the authors note:
Still, in vitro is all well and good, but the team took it a step further and challenged mice with MA10 (mouse-adapted) strains of SARS-CoV-2 D614G and XBB.1.5 variants and examined the protection of these 3 antibodies:
SC27 completely suppresses viral replication in the lungs and prevents weight loss of mice. I do think that the research team could have used k18-hACE2 mice here without needing MA10 virus since the point here was to examine protection, not necessarily model pathogenesis, but this choice on their part is certainly not wrong. It would also have been helpful to see whether mice treated with these antibodies and then housed with other mice were infectious, but the work involved here was already heroic.
In addition to this, the research team profiled the details of why SC27 is so special:
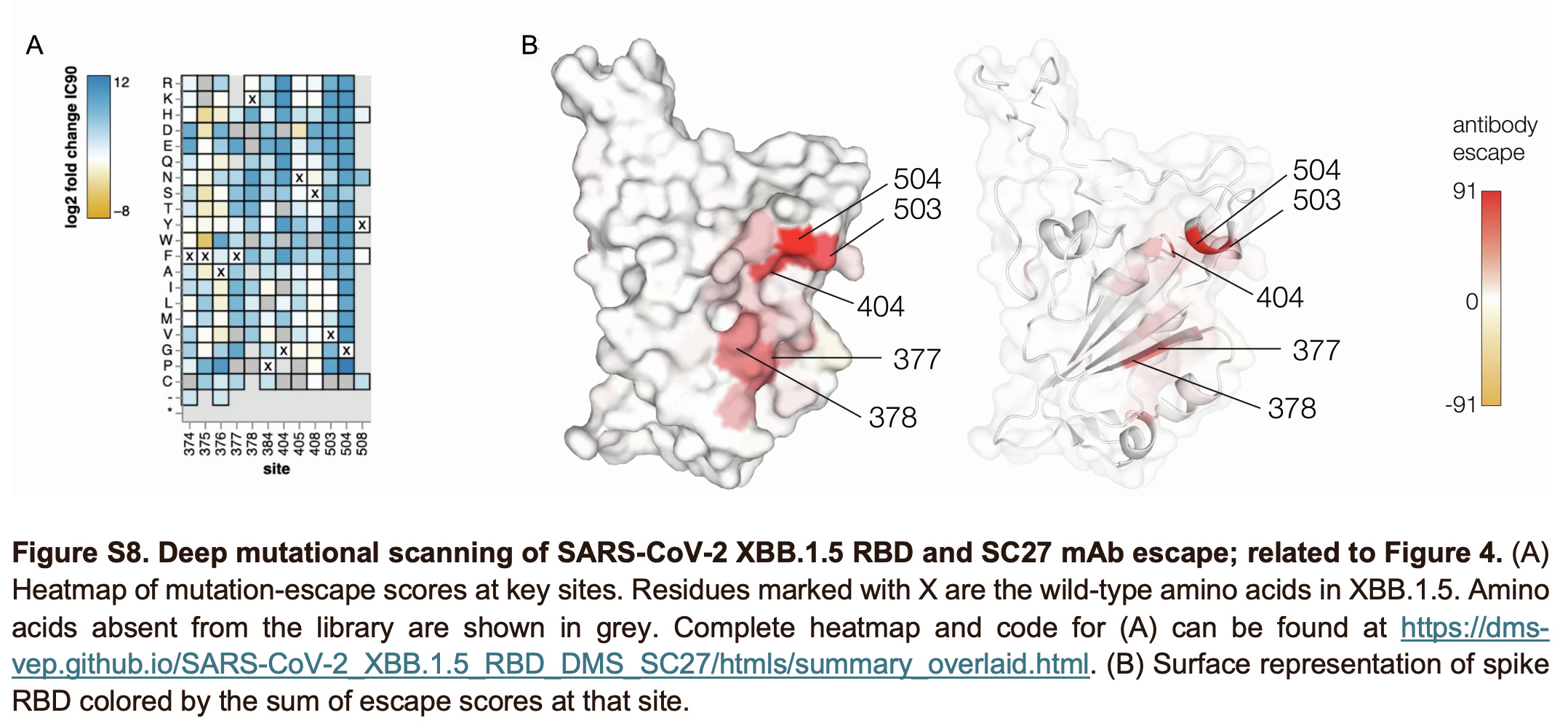
In brief, SC27 manages to target a highly conserved site (which is shared by class 4 antibodies) while also overlapping with the class 1 site (giving it incredible potency- in fact, it is so high that it overturns what was previously believed to be the theoretical limit on how tightly an antibody can bind). Antibodies of this type were previously found exclusively in those who had been infected with SARS-CoV-1 in 2003 and then received SARS-CoV-2 vaccines. This work demonstrates that mRNA vaccines alone could elicit antibodies targeting this site. Furthermore, the team determined what it took for the spike protein to escape SC27. Turns out… a lot. In fact, basically none of the relevant mutations are found in SARS-CoV-2 in circulation even today or at basically any point during the pandemic. Furthermore, even when some mutations did occur (such as at site 504 in the case of Pangolin-GX-2017), the antibody retained potent binding, suggesting that many of these mutations will have to occur together for true escape to occur.
Questions you might have and my best attempt to answer them
If SC27 can be elicited by hybrid immunity and has all these incredible properties, why is SARS-CoV-2 still around being awful when nearly everyone already has hybrid immunity?
This is probably the most important and most obvious question that comes of this. There are a few things to note. Firstly, SC27-LV, the precursor to SC27, wasn’t all that impressive when it came to neutralization breadth or potency. It’s likely that the precursors are there, but the ability to coax SC27-like antibodies might not be as widespread as we hope and might need more support than what current vaccines and infections can offer when it comes to making these kinds of antibodies common in the public. Genetics may also play a role here- the genes encoding antibodies vary across individuals, which can affect the tendency to make particular antibodies. For example, those that had certain variants of IGHV1-2 were found to be unable to make a class of antibodies known as VRC01, which broadly neutralize HIV. However, this is perhaps not as much of a concern when it comes to antibodies specifically because somatic hypermutation should essentially permit any sequence to occur so long as it is not actively selected against (in contrast to T cells, which are constrained by a person’s MHC haplotype and TCR repertoire). It may, however, take coaxing with different antigens depending on a person’s genetic background. Notably, a vaccine for HIV (eOD-GT8 60mer) that reliably induces VRC01 precursors in 97% of humans it was given to in a phase 1 trial has been identified, suggesting that it is possible to overcome the constraints antibody genetic variation places.
SC27-like antibodies are commonly found in those who had SARS-CoV-1 in 2003, which brings me back to my regular refrain for a vaccine incorporating SARS-CoV-1 spike. It’s also worth taking heed that SC27 is but a single antibody being made by a single set of plasma cells derived from the same clone- its concentration within the serum (which contains antibodies from *millions if not billions* of plasma cells) is not going to be high. We have not yet made a deliberate effort to try eliciting these class 1/4 antibodies with vaccination. This suggests… we maybe really should.
It’s also worth noting that SC27 came from a person receiving 2 doses of vaccine and then experienced a breakthrough infection. 3 doses of mRNA vaccine have previously been shown to produce antibody responses similar to this, but the exact prevalence of SC27-like antibodies (both within individuals and across the population) is not entirely clear. It has also been reported that the third dose of mRNA vaccine preferentially elicits class 1/2 antibodies whereas in infection, more of the antibodies are class 3.Can we just give SC27 to people?
After modification of the antibody for desired properties (e.g., prolonged half-life, desired Fc effector functions), an appropriate clinical trial process establishes its safety and effectiveness, and regulators review it and grant approval, yes. Per the paper, a patent for the antibody is still being worked out but after that and appropriate commercial partners are found, it should be feasible that those at high-risk might be able to take the antibody similar to what is done for nirsevimab for RSV, or pemivibart and (formerly) tixagevimab/cilgavimab for COVID-19. For everyone else… that’s an open question. With nirsevimab, there is a very broad recommendation in the US for very young children (all kids under 8 months and some older kids with additional risk factors) and we ran into significant supply issues initially with that, so the prospect of a monoclonal antibody given to everybody (not just babies) here is not totally clear in terms of feasibility and cost.
There is another potential consideration here though if we do opt to go that route: if we were to vaccinate with a spike protein containing the specific site that SC27 binds, it would block our ability to elicit antibodies our own immune system makes against that site (this is known as epitope masking and it helps to diversify the immune response). So long as we have SC27, that shouldn’t be an issue, but, it means we won’t make our own antibodies that bind sites that overlap with the footprint of SC27, so long as SC27 concentrations remain high.Can SARS-CoV-2 (or other SARS-like coronaviruses) evolve to escape SC27?
With coronaviruses, never say never. However, the excellent conservation of the SC27 footprint across so many different SARS-like coronaviruses and the ability of the antibody to tolerate mutations to key amino acid residues for binding is definitely encouraging. At the same time, the common cold coronavirus NL63 also uses ACE2 despite being an alphacoronavirus (and therefore not SARS-like), and per this paper, NL63 is not neutralized by SC27. Antibodies that can neutralize NL63 and SARS-CoV-2 (as well as other coronaviruses) have been described, but these target S2, which is well-conserved but the antibodies require high concentrations to be neutralizing (levels can be lower if C1q is also present). It may be important to see whether RBD antibodies that cross-neutralize NL63 and SARS-like coronaviruses could be induced in the pursuit of a universal vaccine, although it should be noted that at the sequence level, similarity in the spikes is minimal (HCoV-NL63 S protein shares only 25% and 17.1% of aa sequence identity with SARS-CoV and SARS-CoV-2, respectively)5.
This is a joke.
Strictly speaking, the part of the spike protein that binds ACE2 is known as the receptor-binding site (RBS) aka receptor-binding motif (RBM) which is located within the RBD. The image below helps clarify:
A is a cartoon with the domains labeled if the spike protein were rolled out into a linear polymer. B shows the specific sequences that define parts of the spike protein with elements of secondary structure labeled. C shows how the spike protein actually looks engaging with ACE2 once it’s folded with relevant amino acid residues and regions (e.g., RBM) labeled.
The class 1/2/3/4 classification of RBD antibodies seems to be the most common one used in the field, but multiple schemes exist and there is no “official” one.
Dejnirattisai and colleagues likened the RBD to a torso and defined epitopes based on analogy to the anatomy of the torso:
This defines 5 groups of epitopes: left shoulder, neck, right shoulder, right flank, and left flank.
Another classification system from Cao and colleagues defines epitope groups A-F and is also occasionally seen. These were defined through unsupervised clustering after profiling antibodies’ behavior in response to escape mutations. To summarize:
Class A and B antibodies can bind to the RBD only in the “up” conformation and are represented by LY-CoV016 (etesevimab) and AZD8895 (tixagevimab), respectively
Class C and D antibodies can recognize the RBD in either the up or down configuration and are represented by LY-CoV555 (bamlanivimab) and REGN-10987 (Imdevimab)
Group E and F target the S309 (sotrovimab parent antibody) site and CR3022 site, which could exhibit pan-sarbecovirus neutralization capacity; these usually neutralize by mechanisms other than interference with ACE2 engagement.
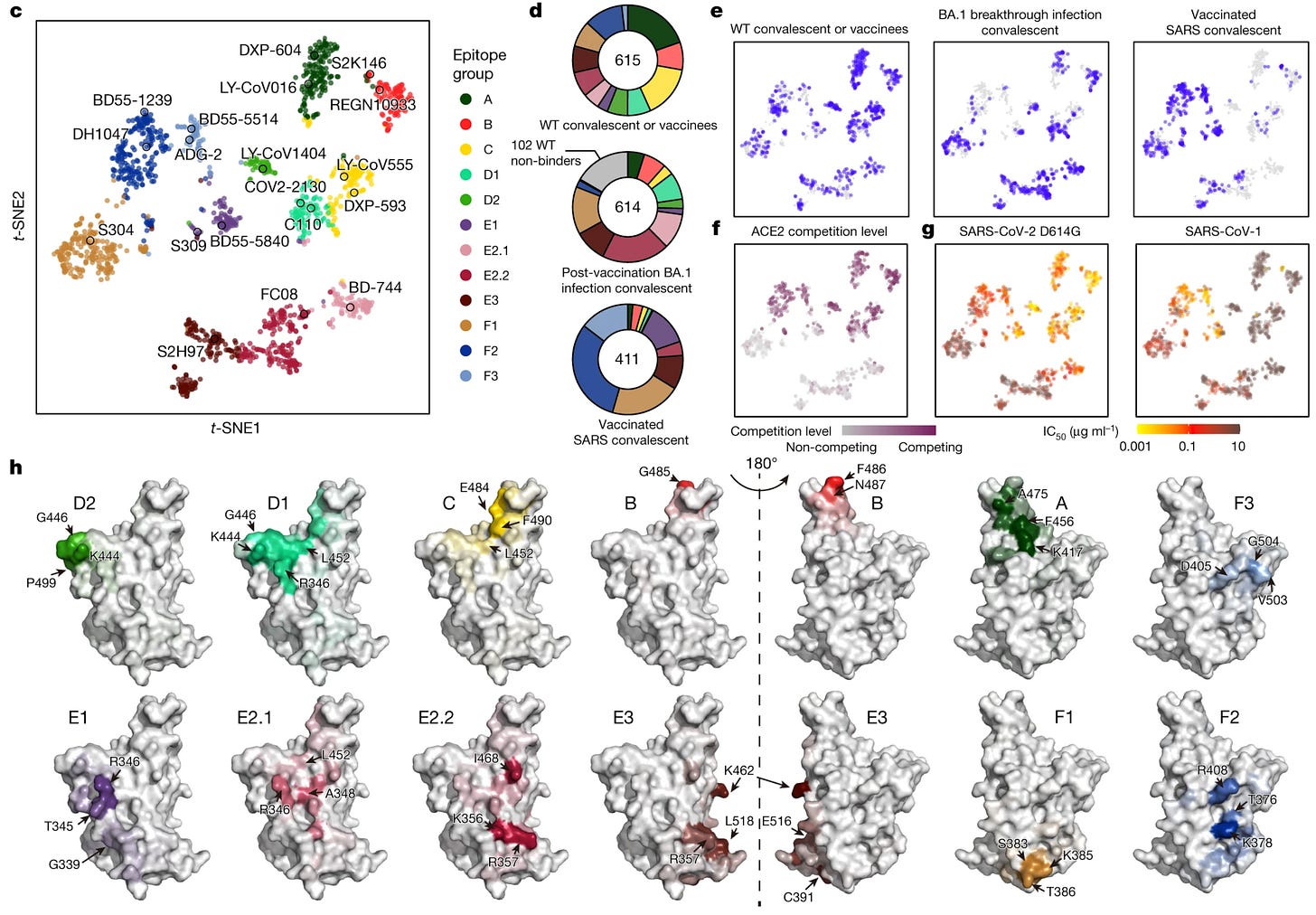
In an update, Cao and colleagues report 12 epitope groups based on deep mutational scanning of 1,538 antibodies. This is a technique in which the antibody target undergoes mutations to establish which sites are important for the antibody to recognize the target:
These groups are A, B, C, D1, D2, E1, E2.1, E2.2, E3, F1, F2, and F3. Mostly, these are the same classes as those in the previous taxonomy, but defined more granularly based on what mutations affect their epitopes:
A-C are the same as those of their previous taxonomy.
Group D antibodies bind to the linear epitope 440–449 on the RBD and are divided into D1 and D2 subgroups. Group D1 is more affected by mutations of R346 and L452, whereas D2 antibodies are not and interact more with P499
Group E1 occupies the S309 (sotrovimab parent) binding site, whose epitope involves G339, T345 and R346.
Group E2 antibodies bind to the front chest of RBD (based on the classification scheme of Dejnirattisai and colleagues). Group E2.1 binding is most affected by mutations of R346 and A348, whereas E2.2 is most affected by K356 and R357.
Groups E3 (S2H97 site) and F1 (S304 site) bind to highly conserved regions on the bottom of the RBD, interacting mainly with K462/E516/L518 and S383/T385/K386, respectively
F2 and F3 antibodies compete with ACE2 and their binding is affected by T376, K378, D405, R408 and G504, corresponding to class 1/4
Another scheme, used by Hastie and colleagues, classified the binding sites through competitive surface plasmon resonance experiments:
Rao and colleagues summarize:
RBD-1, RBD-2, and RBD-3 class antibodies bind to the receptor binding motif of the RBD at different angles, and the “up” RBD conformation is required for the binding. RBD-4 and RBD-5 class antibodies bind to different positions on the outer surface of the RBD when it is in the “up” or “down” state. For both RBD-6 and RBD-7 class antibodies, they bind to the inner surface of the RBD, which is often referred to as a cryptic epitope, and two “up” RBDs are necessary.
Rao and colleagues also go on to define a cryptic epitope known as RBD-8. Antibodies targeting RBD-8 can rescue the function of antibodies targeting RBD-5 when variants evolve resistance.
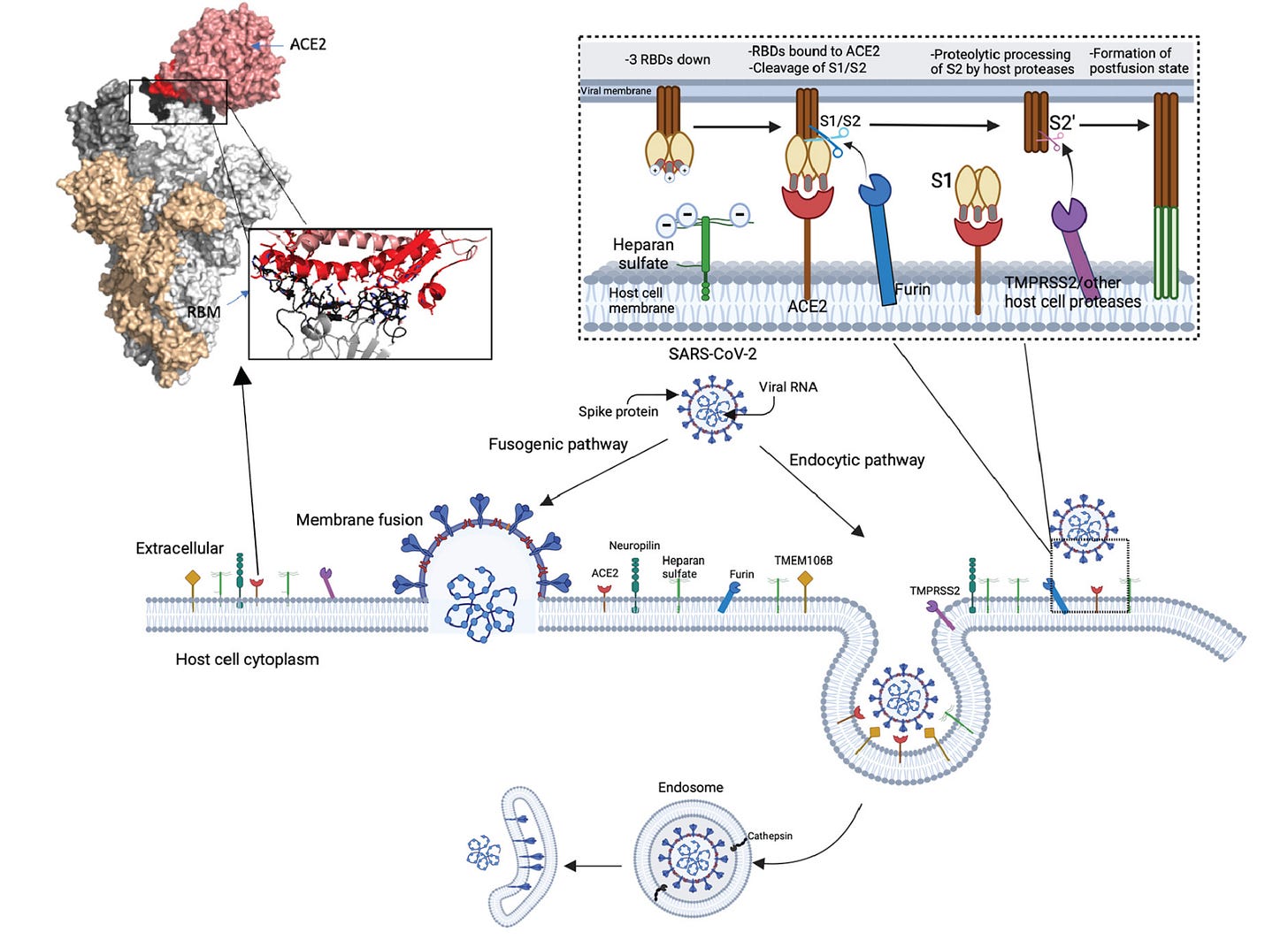
I thought it might be helpful to explain a bit about the structure of spike protein here, but it’s not absolutely required. SARS-CoV-2 viral particles each have 24-30 or so spike proteins on their surface. Each spike on the surface is a homotrimer, meaning it comprises 3 identical spike proteins which associate with one another. As the spike protein binds its targets, it can change conformations (i.e., shape). The receptor-binding domain conformation can be described as being either “up” or “down” as shown below:



These might also be known as “open” or “closed” respectively. When SARS-CoV-2 engages with ACE2, one of the spike proteins must transition from the down (closed) state to the up (open) state to expose the receptor-binding motif (RBM). When this happens, proteases from the host (e.g., TMPRSS2) cleave the spike protein at a site called S2’, which exposes the fusion peptide that allows the coronavirus to fuse its membrane with that of the host cell, releasing its genome into that cell and beginning its replication. In general, this occurs at the cell membrane, but depending on the cell type and what proteases it has on the surface, SARS-CoV-2 could be taken up into an endosome and its spike protein can be cleaved by proteases within that endosome:
It’s worth mentioning that in addition to directly mutating the key epitopes of the spike protein that neutralizing antibodies recognized, Omicron’s spike protein underwent additional changes that make it more effective at evading antibodies:

For example, the Omicron spike protein is much more tightly packed than pre-Omicron variants. In addition, Omicron’s spike has increasingly evolved to be predominantly in the down conformation, which allows it to evade antibodies from class 1 and class 4. Class 2 and 3 antibodies can recognize the down conformation, but often lack breadth, which makes them relatively easy to escape by antigenic drift. This change also allows it to evade certain antibodies which have a quaternary binding mode, i.e., bind multiple spike proteins at the same time to work. In principle, this makes it harder to engage with ACE2 as RBD must be in the up conformation to do that, but in the setting of high levels of neutralizing antibodies, this would be advantageous. Notably, while Omicron initially seemed to prefer the 2-down, 1-up conformation, XBB.1.5, EG.5, and BA.2.86 have all been reported as being predominantly in the 3-down conformation, which allows for even more extreme evasion of the immune system, but at the cost of ACE2 binding.
While, in general, antibodies can be diversified solely through class switch recombination and somatic hypermutation, in exceptional cases, some additional mechanisms have been seen. For example, LAIR1 (leukocyte-associated immunoglobulin-like receptor 1, or CD305) and LILRB1 (leukocyte immunoglobulin-like receptor 1 aka CD85J) bind to proteins expressed by the malaria parasite Plasmodium falciparum known as RIFINs (repetitive interspersed families of polypeptides). It turns out that in malaria, antibodies can copy large stretches of these receptors directly into their genes, resulting in, essentially, fusion proteins which contain insertions from LAIR1 or LILRB1 in addition to the typical immunoglobulin structure. This is especially incredible because LAIR1 and LILRB1 are located on 19q13.4 in a region known as the leukocyte receptor cluster, whereas the antibody loci are on chromosome 14 in the case of the heavy chain, and light chain located on chromosome 2 and 22 for κ and λ respectively. These result in broadly reactive antibodies that can neutralize malaria parasites. Shockingly, 5-10% of donors from Tanzania and Mali demonstrate these LAIR1 antibodies, and through somatic hypermutation, these antibodies gain enhanced binding to RIFINs and abolish binding affinity to collagen. European donors also demonstrate very large insertions in the elbow joints of their antibodies but these do not correspond to LAIR1. It has been proposed that this occurs specifically in malaria because the infection induces genetic instability. No such antibodies where there is an ACE2 insertion analogous to the LAIR insertion have been described as naturally occurring with SARS-CoV-2 or other coronaviruses (to the best of my knowledge). We have, however, seen antibodies against ACE2 that do not interfere with its normal function or reduce its expression on cells that do protect from SARS-CoV-2, and antibodies that mimic the surface of ACE2 (class 1 antibodies) have also been reported as being induced through SARS-CoV-2 infection. Though this antibody, S2K146, demonstrated substantial breadth, it was less resilient to changes in its binding footprint that occurred in other sarbecoviruses